Bachelor Thesis: In-silico identification, production and verification of the receptor binding protein of the Paenibacillus phage HB10c2
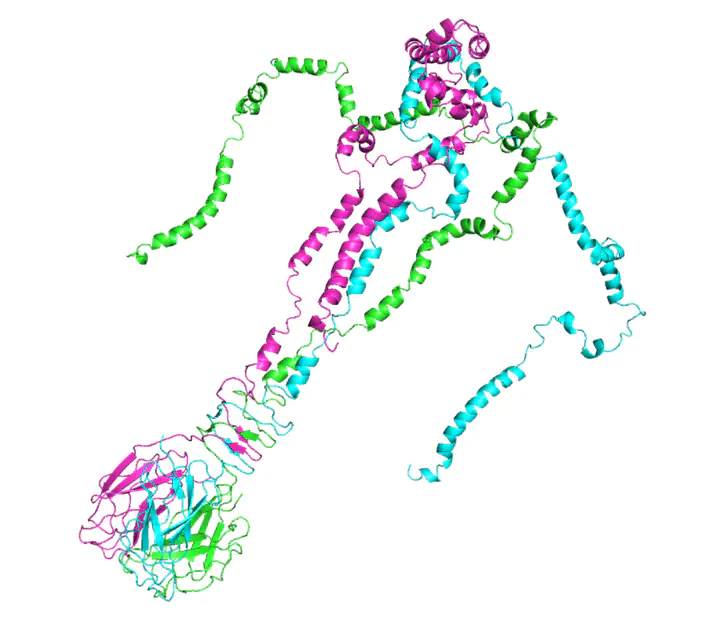 Protein folding prediction of gp17 as trimer by Henrik Seyfried (iGEM team member), the target candidate of the thesis
Protein folding prediction of gp17 as trimer by Henrik Seyfried (iGEM team member), the target candidate of the thesis
Abstract
Receptor binding proteins (RBP) of bacteriophages are involved in the specific recognition and binding of bacteria via all types of receptors. In the phage subclass Siphoviridae they are located at the most distal part of the tail, anchored in the baseplate. The baseplate consists of a varying number of proteins for each phage and is responsible for connecting the phage with the host bacterium to enable the phage to inject its DNA into the cytoplasm. These proteins are key for the binding mechanism of phages, but rather limited information is available in literature. Since the specific recognition of bacterial cells has a great potential for whole cell detection and other applications, it would, however, be important to obtain a better understanding of these proteins (RBPs). In the course of my studies, the gene product gp17 (HB_00017) has been identified as a likely candidate for being the RBP of the Paenibacillus phage HB10c2. The sequenced genome of HB10c2 were compared with the genome of other phages and putative protein candidates were modelled with Phyre2. Subsequently, the protein was produced heterologously in Escherichia coli. However, it seemed to be insoluble. A newly designed test for RBPs based on Ni-NTA affinity chromatography failed to confirm the interaction of gp17 with Paenibacillus larvae. A colony-assay for the identification of RBPs suggested that the protein encoded by HB_00017 could be the RBP of HB10c2.
This thesis was done in parallel with the iGEM-project Beeosensor, which uses the bacteriophage whose RBP is the topic of this publication. The trimer-model of the protein (picture above) was created by my team colleague Henrik Seyfried.